PIKAIA - Breeding in Genetic Algorithms
This Page gives further detail on the breeding procedureincorporated in the genetic algorithm-based optimization subroutinePIKAIA. The discussed is framed in the context of a genericmaximization problem that consists in searching for the (x,y) pairthat maximizes the output of a (given) function f(x,y),in a domain bounded between 0 and 1 in the xand y directions.This defines a two-parameter optimization problem.
1. Encoding and Decoding
Suppose that two parents (i.e., two (x,y) pairs) have beenselected for breeding. The first step is to construct an encodingof each parent, in the form of a string-like structure orchromosome. PIKAIA proceeds by directly fragmenting eachparameter into simple decimal integers, and splicing the resultingintegers strings one behind the other.The number of digits (nd) to be retained in the encodingmust be specified externally. For example, with nd=8 one wouldobtain(x,y)=(0.123456789,0.987654321) -> 1234567898765432
The length of the chromosome is then nd times the number ofparameters (n) defining the solution. Here n=2 so the chromosomehas length 16. Each digit can be thought of as a geneoccupying a chromosomal site for which thereexists 10 possible allelles (possible gene values).Decoding is simply the reverse process:
1234567898765432 -> (0.12345678,0.98765432)=(x,y)
Not that in terms of the original parametervalues, a truncation loss has occurred in running through theencoding/decoding processes.
2. Crossover
The first step of the breeding process proper is the application ofthe crossover operation to the pair of parent chromosomes.Consider now two parent chromosomes produced by the encoding processdescribed in the preceding section:1234567898765432 Parent #17654321023456789 Parent #2Randomly select one of the 16 sites, cut both chromosomes at that site,interchance the fragment right of and including the cutting site,and splice the fragments back: for a cut at site #10, this wouldlook like:
CUT: 123456789 8765432 765432102 3456789There results of this two new offspring chromosomes, eachcontaining intact chunks of chromosomal material from each parent.In the GA literature this is called one-point uniform crossover; ``one point'' because there is only one cutting siteper chromosome pair, ``uniform'' because each site is equally likelyto be selected for the cut/swap/splice operation. In general oneintroduces a probability Pc that the crossover operationis to occur; unlike the mutation probability (see below), Pcshould not be very much smaller than one.
SWAP: 123456789 3456789 765432102 8765432
SPLICE: 1234567893456789 Offspring #1 7654321028765432 Offspring #2
3. Mutation
The second step of the breeding process is the application ofthe mutation operator to each offspring chromosome. For each geneof each offspring chromosome, a random number between 0 and 1is generated, and if this number is smaller than a presetmutation probability Pm then the gene value is randomly changedto any other legal value. For example, the following is a mutationat site 2:7654321028765432 7 54321028765432 7154321028765432
In the GA literature this is called one-point uniform mutation; "one point" because there is only one gene affectedat a time, "uniform" because each gene is equally likelyto be subjected to mutation, independently of the site it occupiesalong the chromosome.
The effects of crossover and mutation on the decoded versionof a parameter set can be drastic (if one of the leading digitsis affected) or quite imperceptible (if one of the least significantdigit is affected). The result is that the breeding process cancause large jumps in parameter space, as well as slight displacements;the resulting search algorithm can explore efficiently, as well as performfine tuning. In the case of the two parameter example used here,we would have gone from two parents:
(x,y)=(0.12345678,0.98765432)
(x,y)=(0.76543210,0.23456789)
to two offsprings:
(x,y)=(0.12345678,0.93456789)
(x,y)=(0.71543210,0.28765432)
Clearly the offsprings have taken sizable jumps throughparameter space, as compared to their parents.
4. Why this crossover business?
The crossover operation is what distinguishes genetic algorithmsfrom other adaptive stochastic techniques, and to a large degreeis responsible for the efficient exploration capabilitiesof genetic algorithms. Consider again one of the 16-gene chromosomedefined previously; replace now each gene by one of three possiblesymbols ("-", "+" or "X") according to the following coding:if the gene value is "good", in the sense of giving its bearer (on average)higher-than-average fitness, replace the gene by the symbol +;if its bearer has below-than-average fitness, by -; if the genevalue does not seem to matter, by X (this would usually be the case forgene encoding least significant digits early in an evolutionaryrun). Here's two possible result of this process for a 16-genechromosome:++--XXXX---+XXXX Parent #1---++XXX+++XXXXX Parent #2Crossover occurring at site #9 (without any subsequent mutation)would produce
++--XXXX+++XXXXX Offspring #1---++XXX---+XXXX Offspring #2Clearly, Offspring #1 is doing a lot better than any of its parents(having 1's in its sites decoding to the first two significant digits),while Offspring #2 will be mostly below average fitness, and so willhave a low probability of being selected for breeding at the nextgenerational iteration. Observe that crossover can combine advantageous"chunks" of "good" chromosomal material in a single offspring.With natural selection operating,these chunks will end up (on average) being copied into thenext generation rather frequently, the more so the more the luckyoffspring finds itself above average. This is the essence of theschema theorem, which forms the basisof genetic algorithm theory.
Technical details:
Consider the set of "good" (i.e., coded "+") genes ofOffspring #1 of the preceding discussion:++......+++.....where the period symbol is used to fill sites that are notpart of the set of genes.This defines a schema, which we will denoteby s. The order of the schema (denotedas N(s)) is the number of participating sites (N(s)=5 here),and its defining length (denoted D(s)) is the lineardistance (measured in sites) between the first and last ofits defining components (D(s)=11 here;the full chromosome length L is 16). Denote now bym(s,t) the fraction of population members containingthe schema s in their chromosomes at generation t, letf(s) be their average fitness, and and F bethe average fitness of the population as a whole.With natural selection operating, it can be shownthat m will vary from one generation to the next as
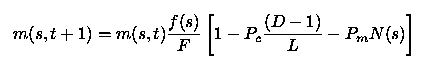
This (discrete) equation expresses the fact that the frequencyof occurrence of favorableschemata (meaning schemata for which f(s)/F is largerthan one) will increase from one generationto the next. If the average fitness were to remain constant(which it won't), that growth would be geometrical. In practicethe growth will not be geometrical, but it will be fast; crossoverleads to the grouping of advantageous genetic chunks, originallydistributed throughout the gene pool. The quantity within thesquare parenthesis refers to the probability of a given schemabeing disrupted/destroyed by the crossover and mutationoperation, a topic explored at greater depthin the question section below. Note that it differs a bitfrom the similar expression given in chapter 2 ofGoldberg's book, due to the slightly different definitionof the crossover operator adopted above.
Questions and Answers:
- For those having already worked their way throughthe selection subPage; what is the most important way in whichthe breeding procedure defined above differs from that usedin the WEASEL example ?[answer]
- Under the assumption of zero crossover and mutationprobability, and with a fixed average fitness Ffor the population as a whole, can you obtain a (discrete)equation expressing the growth in the frequency m(s,t)of a schema whose carriers' average fitness f(s) exceeds theaverage by a fraction c, i.e., f(s)=F+cF ?[answer]
- Can you construct general mathematical expressions for theprobability of a schema of a given order(N) and defining length (D) not being disruptedby (a) the crossover operator, (b) the crossover operator,(c) either the crossover or mutation operator. Assume afull chromosome length equal to L.(d) How does your result in (c) compare to the quantitywithin square parentheses in the expression cited above ?Can you explain the difference ?[answer]
- Based on your answers to question 3, what are the optimaldefining characteristics of a schema that will minimizedisruption upon breeding ?[answer]
