If you are interested in using the genetic algorithm based FORTRAN-77 optimization subroutine PIKAIA, and the modeling fitness-function that you want to maximize is computationally intensive, you have come to the right place. Here you will find all that you need to run PIKAIA in parallel on the supercomputers at the National Center for Atmospheric Research.
MPIKAIA is a slightly modified version of the PIKAIA 1.2 code developed by Paul Charbonneau and Barry Knapp at the High Altitude Observatory. It uses MPI code to parallelize the "Full Generational Replacement" evolutionary strategy. This package comes with a sample fitness function (userff.f), which you should replace with your own function/model after you successfully compile and run the sample code. The code has been verified to compile and run properly on several NCAR supercomputers (frost, lightning, bluesky). It also will run on multi-processor Linux machines, if they have MPI installed and configured properly.
If you use MPIKAIA and publish the results, please consider citing the paper that describes this code (Metcalfe & Charbonneau, 2003, Journal of Computational Physics, vol. 185, pp. 176-193). A freely accessible preprint version of this article is available at arXiv:astro-ph/0208315.
cd mpikaia/src/
gmake
The Makefile should automatically determine which architecture you are compiling on, and set the options appropriately. The default build is mpikaia which will be placed in the mpikaia/ directory.
cd mpikaia/
runjob.csh <seed_int>
The runjob.csh script should automatically determine which machine you are running on, and submit a test job through the local batch queue system (<seed_int> is an integer to seed the random number generator).
In addition to decomposing the function of the code, a further division based on the data was also possible. Fitness evaluation across the population is inherently a parallel process, since each model can be evaluated independently of the others. Moreover, it requires minimal transfer of information, since all that the user-supplied function requires is the n-dimensional floating-point array of parameters defining a single instance of the model, and all it needs to return is the floating-point value corresponding to the model's fitness. It is then natural to send one model to each available processor, so the number of machines available would control the number of models that could be calculated in parallel. Maximal use of each processor is then assured by choosing a population size that is an integer multiple of the number of available processors.
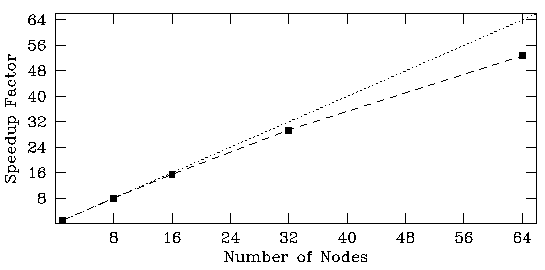
In practice, this recipe for dividing the workload between the available processors proved to be very scalable. Since very little data is exchanged between the master and slave tasks, our 64-node Linux cluster provided a speedup factor of about 53 over the performance on a single processor.
Master Subroutine
Starting with the source code for PIKAIA 1.2, we used the message
passing routines of MPI to create a parallel fitness evaluation subroutine.
The original code evaluates the fitnesses of the population one at a time
in a DO loop. We replaced this procedure with a single call to a new
subroutine that evaluates the fitnesses in parallel on all available
processors. This parallel version of PIKAIA
(pikaia_master.f) along with the parallel
fitness evaluation subroutine (mpi_fitness.f)
constitutes the master program.
After starting the slave program on every available processor, mpi_fitness.f sends an array containing scaled values of the parameters to each slave job over the network. In the first generation, these values are completely random; in subsequent generations, they are the result of the selection, crossover, and mutation of the previous generation, performed by the non-parallel portions of PIKAIA. Next, the subroutine listens for responses from the network and sends a new set of parameters to each slave job as it finishes the previous calculation. When every set of parameters in the generation have been assigned a fitness value, the subroutine returns to the main program to perform the genetic operations, resulting in a new generation of models to calculate. The process continues for a fixed number of generations, chosen to maximize the success rate of the search.
Slave Subroutine
To allow many instances of our code to run simultaneously, we added a
front end that communicates with the master program through MPI routines.
This code (ff_slave.f) combined with the
user-defined fitness function (userff.f)
constitutes the slave program, which runs on every available processor.
The operation of each slave task is relatively simple. Once it is
started, it waits to receive a set of parameters from the master task
over the network. It then calls the fitness function (the sample fitness
function twod in this distribution) with these parameters as
arguments, and returns a fitness value to the master task.
Compiling and Running
This version of MPIKAIA has been compiled and tested
under the MPI distributions available on NCAR supercomputers, and
with MPICH 1.2.5
running on a multi-processor Linux system. You can compile the sample
program using gmake with the provided
Makefile. The code can then be run in parallel
with the runjob.csh script, by issuing a command
like: runjob.csh <seed_int> (the command line argument is
an integer to seed the random number generator).
Production runs may also need to modify some of the parameters that control the batch queues in the runjob.csh script, like the number of processors to use and the estimated runtime. To run with maximum efficiency, the number of processors should be set to an integer fraction of ctrl(1), plus 1 for the master program.